Lab Projects
The overarching research theme of this lab is investigating the genetic underpinnings of brain connectivity at the circuit level and the behavioral and pathological phenotypes they present in neuropsychiatric disorders. We will investigate the structural and functional connectivity of the brain on a macro-scale using clinical neuroimaging datasets and on a micro-scale using single-neuron resolution imaging and genomics datasets collected from small animal model systems. Deciphering the genetic correlates of brain connectivity in health and disease will have a broader impact in basic neuroscience and clinical research to better understand neuropathogenesis and enable the design of precision medicine treatments. PDF document with project details can be found here. Slide deck that covers broad lab policies is found here.
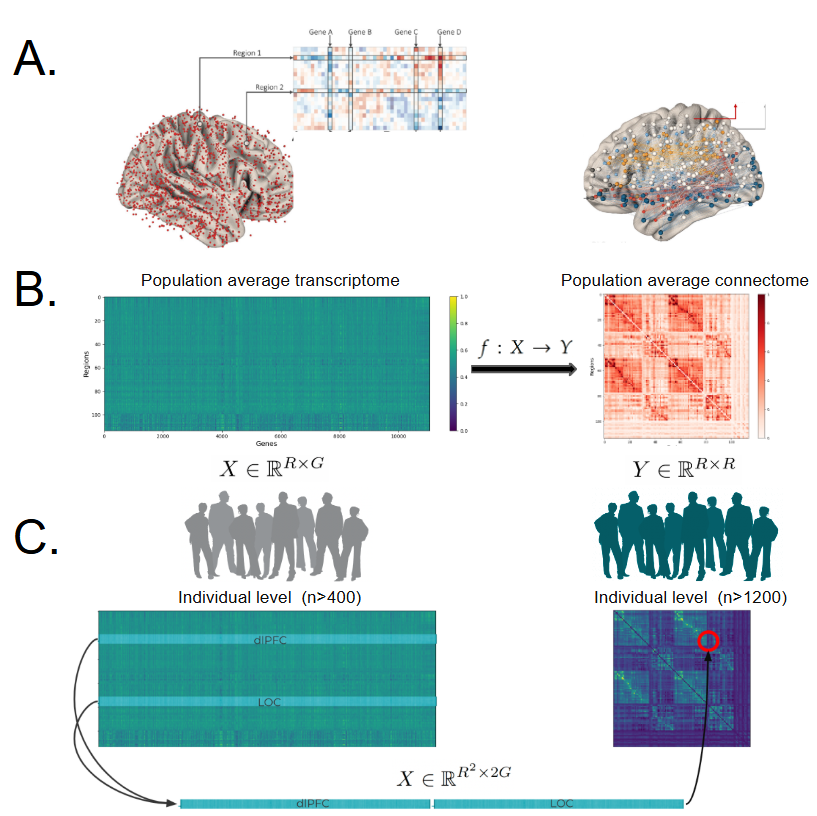
Spatiotemporal Integration of the Human Transcriptome and Connectome
A longstanding goal of systems neuroscience is to understand the molecular landscape underlying macro-scale brain structure and function. With increasing availability of diverse human neuroimaging and genomic datasets, there is now a unique opportunity to develop robust computational models to predict how gene expression influences neural connectivity across the adult lifespan and among clinical populations.
Collaborators: Junhao Wen, Christos Davatzikos
Genetic basis of the fruit fly connectome
We will use single-cell RNA sequencing data together with maps of brain connections from the fruit fly to study how gene activity relates to the way neurons are connected. Our goal is to create a computer-based method that can help predict and understand how synapses form and function.
Collaborators: Richard Mann, Himanshu Gupta
Connectivity and coactivity predicts cell-type in mouse hippocampus
Using paired transcriptional information and calcium activity, can we link cell type identity with connectivity information?
Collaborators: Losonczy Lab
Seq2Connectome
Our goal is to predict the connectome of C. elegans from DNA sequence data. Because C. elegans is so well studied, we have a rich dataset upon which to build a computational model, which we hope to generalize to other organisms.
Collaborators: Arman Afrasiyabi, Alexis Weinreb
Disease gene classification using multi-species gene interaction network data
Our goal is to classify genes(represented as nodes in the graph) into disease association or non-disease association. There are set of nodes which are non-omim genes. By having done this we can study other genes which are at greater risk of being potentially associated with disease which are under studied and hence marked as non-disease genes.
Collaborators: Sandeep Wontakal
Multi-species blind localization of brain regions using local field potentials
In electrophysiology, precise in vivo localization of recording sites in deep brain structures is crucial for consistent targeting in multi-day recordings and accurate deep brain stimulation while current approaches are limited by either spatial or temporal resolution. Here we develop a learning-based automatic localizer to identify mice hippocampus sublayers from high-density extracellular recordings and apply our model to human chronic recordings.
Collaborators: Buzsaki Lab (Anna Maslarova, Mihály Vöröslakos), Arman Afrasiyabi, Cole Hurwitz
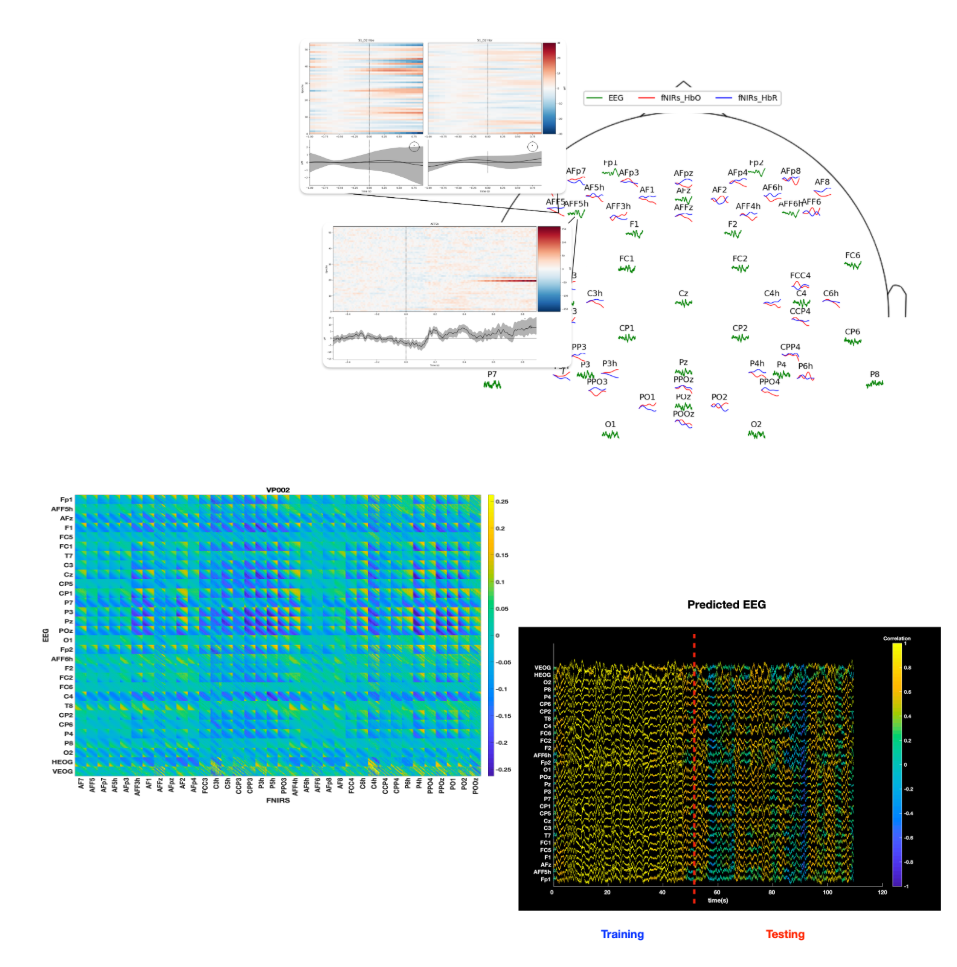
Transcoding of voltage signal and hemodynamic response in human neuroimaging
Mobile non-invasive neuroimaging technologies such as electroencephalography (EEG) and functional Near-Infrared Spectroscopy (fNIRS) facilitate the recording of neural states in naturalistic settings. However, these modalities face significant resolution limitations. This work aims to denoise and up-resolve non-invasive BCI developing modality transcoding techniques of simultaneous EEG and fNIRS recordings.
Collaborators: Arman Afrasiyabi, Cole Hurwitz
Multimodal registration of in-vivo vs. ex-vivo cell populations using calcium imaging and spatial transcriptomics
The registration problem aims for precise mapping of cells integrating imaging from different modalities. The challenge arises from differences in scale, resolution, stability, tissue shrinkage and imaging at different angles in the brain.
Collaborators: Losonczy Lab
Voltage imaging event detection and clustering at subcellular resolution
Can we detect slow and fast spiking events at dendrites using voltage imaging?
Collaborators: Losonczy Lab
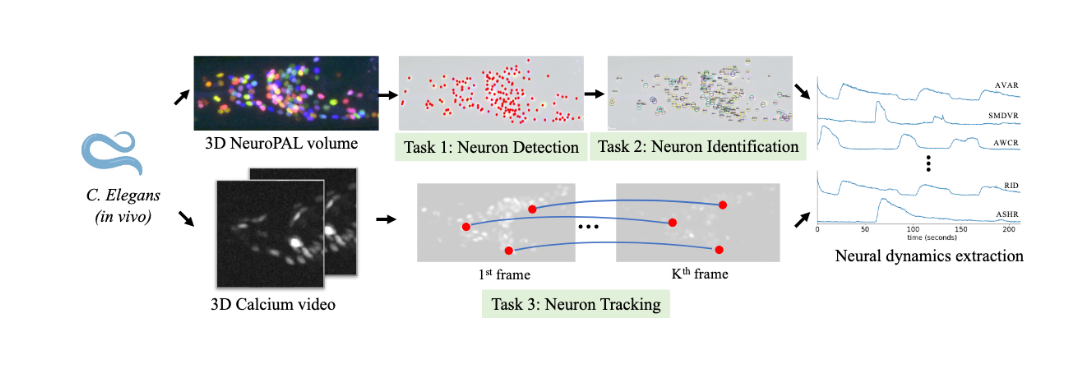
Whole-brain single-cell detection, identification and tracking of neurons in c. elegans during behavior
Can we develop high-throughput and robust tools to track, identify and extract the calcium activity for each neuron in the worm nervous system during behavior?
Collaborators: Donglai Wei
Connectome constrained Spiking Neural Networks
Can we train spiking neural networks whose connectivity or cell type specific intrinsic parameters are constrained by biological networks such as drosophila or mouse cortex? If we can show that connectome constrained SNN’s can be trained such that they faithfully recreate the internal dynamics of their target networks (e.g. a biological neural network), then we can treat the SNNs as a in-silico surrogate of a biological neural network in which perturbation experiments and other phenomena can be studied.